The Gore lab uses experimentally tractable laboratory microcosms to explore how interactions between individuals drives the evolution and ecology of communities.
Three primary areas drive research in the group:
- How do populations behave near "tipping points" leading to collapse?
- How can cooperative behaviors evolve within a species or community?
- What determines the diversity in communities?
What conditions favor the evolution of cooperative behaviors?
We use microbes such as yeast and bacteria to study how evolution can favor cooperative behaviors. As a postdoctoral fellow in the van Oudenaarden Lab Jeff developed sucrose metabolism in yeast as a model cooperative system (Gore et al, Nature (2009)). In this study, we found that cooperators have preferential access to the sugar that they create, thus allowing the cooperators to survive even when in the presence of cheaters. Since that original paper, we have used this as a model system in many other papers. We have also studied how the cooperative breakdown of starch can lead to spatial patchiness (Ratzke and Gore, Nature Microbiology (2016)), and how a cross-feeding mutualism can become parasitic as the environment improves (Hoek et al, PLOS Biology (2016)).
What determines the diversity of multi-species microbial communities?
Microbial communities display remarkable diversity, and this diversity plays a key role in the functioning of the community. A major focus of research in the group is directed towards developing a bottom-up understanding of how these multi-species communities form and function. For example, we are studying the degree to which pairwise competitive outcomes allows for the prediction of survival when bacterial species are placed in competition with a larger number of species (Friedman et al, Nature Ecol & Evol (2017)). We have also found stochastic effects associated with colonization can drive heterogeneity in the worm microbiome (Vega and Gore, PLOS Biology (2017)).
What does the cooperative nature of antibiotic resistance mean for the evolution of resistance? Bacterial resistance to penicillin and related antibiotics is obtained by expressing a protein that degrades the antibiotic, thus removing a public bad. We found that this cooperative behavior is susceptible to "cheating" (Yurtsev, Chao, et al, Molecular Systems Biology (2013)), and also that it is possible for resistant bacteria to form a mutualism to survive multi-drug environments (Yurtsev*, Conwill*, et al, PNAS (2016)). We are also exploring the consequences of this cooperative behavior for the evolution of increased resistance (Artemova et al, MSB (2015)).
Gore Laboratory
Department of Physics
Massachusetts Institute of Technology
Physics of Living Systems Group
400 Technology Square, NE46-602
Cambridge, MA 02139
Department of Physics
Massachusetts Institute of Technology
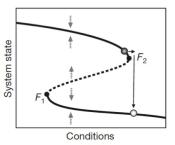
What are the dynamics before the collapse of a population?
Theory from nonlinear dynamics argues that there are characteristic behaviors of complex systems before "tipping points". We have made a significant effort in exploring these signatures of critical slowing down in a number of different experimental systems. We first demonstrated that changes in the fluctuations of a population can be used to anticipate an impending collapse (Dai et al, Science (2012); Chen et al, Nature Comm (2013); Dai et al, PNAS (2016)). Next we showed that there are spatial patterns that emerge before collapse (Dai et al, Nature (2013)). Since then we have extended these ideas to cell decision-making (Axelrod et al, eLife (2015)) as well as intertidal communities at field sites on islands in Italy (Lindi et al, Nature Ecol & Evol (2017)).
GORElaboratory
Ecological Systems Biology